Support Vector Machines for Biomarkers Detection in in vitro and in vivo Experiments of Organochlorines Exposure
Máquinas de vectores de soporte para detección de biomarcadores en experimentos in vitro e in vivo de exposición a organoclorados
Received: July 21, 2021
Accepted: November 22, 2021
Available: December 15, 2021
J. A. Lopera-Rodríguez; M. Zuluaga; J. A. Jaramillo-Garzón, “Support Vector Machines for Biomarkers Detection in in vitro and in vivo Experiments of Organochlorines Exposure”, TecnoLógicas, vol. 24, nro. 52, e2088, 2021. https://doi.org/10.22430/22565337.2088
Highlights
Abstract
Metabolomic studies generate large amounts of data, whose complexity increases if they are derived from in vivo experiments. As a result, analysis methods highly used in metabolomics, such as Partial Least Squares Discriminant Analysis (PLS-DA), can have particular difficulties with this type of data. However, there is evidence that indicates that Support Vector Machines (SVMs) can better deal with complex data. On the other hand, chronic exposure to organochlorines is a public health problem. It has been associated with diseases such as cancer. Therefore, its identification is relevant to reduce their impact on human health. This study explores the performance of SVMs in classifying metabolic profiles and identifying relevant metabolites in studies of exposure to organochlorines. For this purpose, two experiments were conducted: in the first one, organochlorine exposure was evaluated in HepG2 cells; and, in the second one, it was evaluated in serum samples of agricultural workers exposed to pesticides. The performance of SVMs was compared with that of PLS-DA. Four kernel functions were assessed in SVMs, and the accuracy of both methods was evaluated using a k-fold cross-validation test. In order to identify the most relevant metabolites, Recursive Feature Elimination (RFE) was used in SVMs and Variable Importance in Projection (VIP) in PLS-DA. The results show that SVMs exhibit a higher percentage of accuracy with fewer training samples and better performance in classifying the samples from the exposed agricultural workers. Finally, a workflow based on SVMs for the identification of biomarkers in samples with high biological complexity is proposed.
Keywords: Organochlorines, Pesticides, Recursive feature elimination, Multivariate statistical methods, Support vector machines, Metabolomics.
Resumen
Los estudios en metabolómica generan gran cantidad de datos cuya complejidad aumenta si surgen de experimentos in vivo. A pesar de esto, métodos ampliamente usados en metabolómica como el análisis discriminante por mínimos cuadrados parciales (PLS-DA) tienen dificultades con este tipo de datos, sin embargo, hay evidencia que las máquinas de vectores de soporte (SVM) pueden tener un mejor desempeño. Por otro lado, la exposición crónica a organoclorados es un problema de salud pública. Esta se asocia a enfermedades como el cáncer. Identificarla exposición es relevante para disminuir su impacto. Este estudio tuvo como objetivo explorar el rendimiento de las SVM en la clasificación de perfiles metabolómicos e identificación de metabolitos relevantes en estudios de exposición a organoclorados. Se realizaron dos experimentos: primero se evaluó la exposición a organoclorados en células HepG2. Luego, se evaluó la exposición a pesticidas en muestras de suero de trabajadores agrícolas. El rendimiento de las SVM se comparó con PLS-DA. Se evaluaron cuatro funciones kernel en SVM y la precisión de ambos métodos se evaluó mediante prueba de validación cruzada k-fold. Para identificar los metabolitos relevantes, se utilizó eliminación recursiva de características (RFE) en SVM y la proyección de importancia de variables (VIP) se usó en PLS-DA. Los resultados mostraron que las SVM tuvieron mayor precisión en la clasificación de los trabajadores agrícolas expuestos usando menos muestras de entrenamiento. Se propone un flujo de trabajo basado en SVM que permita la identificación de biomarcadores en muestras con alta complejidad biológica.
Palabras clave: Organoclorados, Eliminación Recursiva de Características, Estadística Multivariada, Máquinas de Vectores de Soporte, Metabolómica.
1. INTRODUCTION
Modern analytical technologies such as mass spectrometry, nuclear magnetic resonance, and tandem mass spectrometry facilitate the study of the metabolome. Metabolomics is defined as the quantitative and comprehensive study of metabolites in a biological system [
The complexity of metabolic profiles depends on the conditions in which the data are generated. For example, metabolic profiles from in vitro experiments show low variability, while those from in vivo studies (e.g., with humans) might be highly variable between individuals. This complexity affects the ability of statistical algorithms to make accurate predictions based on metabolic profiles.
Methods such as Principal Component Analysis (PCA), Partial Least Squares Discriminant Analysis (PLS-DA), and Orthogonal PLS-DA (OPLS-DA) are commonly used to analyze metabolomics data. However, some studies have identified that their classification capacity can be suboptimal in studies with real life conditions where several variables cannot be controlled and the data can have a nonlinear distribution [
Support Vector Machines (SVMs) area supervised learning method that generates a model able to map a training dataset with two categories into a higher-dimensional space in order to separate them by a margin as large as possible [
Recent studies have also compared PLS-DA with other methods, including SVMs. Mendez et al. [
Like in PLS-DA, a list of the most relevant metabolites can be generated by SVMs using SVM-Recursive Feature Elimination (SVM-RFE) [
On the other hand, organochlorines are a group of pesticides used to control plagues [
Thus, the aim of this study was to describe the discriminant ability of SVMs to handle samples from both in vitro and in vivo studies and compare their results with those obtained with PLS-DA. In addition, the capacity of SVMs to propose metabolites as candidate biomarkers in the context of organochlorine pesticide exposure was explored.
2. METHODS
2.1. Sample preparation -in vitro study
A secondary dataset from a study published in 2016 [
Subsequently, 36 samples of HepG2 cells were exposed to the organochlorine solutions (i.e., 100 µM of endosulfan, 100 µM of lindane, 50 µM of DDT, and 150 µM of aldrin), a mixture treatment at equimolar concentration, and the controls with DMSO (1 % v/v); six samples per treatment. In addition, the cells were incubated for 24 hours with 5 % CO2 at 37 °C. After such period of exposure, cellular metabolism was inactivated, and endogenous metabolites were extracted adopting the quenching methodology previously published in [
The information was processed as follows. First, the signals were automatically deconvolved using ChromaTOF software. Then, the data were extracted without smoothing, and peaks were detected at signal/noise ratios of 5:1 and a peak width of 3 s [
Finally, 1081 signals were deconvolved. Those with more than 30 % missing values were discarded, leaving 399 signals related to potential metabolites, out of which 153 were identified and 246 remained unidentified. The dataset obtained was composed of 6 classes (aldrin, DDT, endosulfan, lindane, mixture, and DMSO) and 153 features.
2.2. Sample preparation -in vivo study
A secondary dataset from a study into agricultural workers exposed to different pesticides was used here. In that study, plasma samples were collected from 100 agricultural workers on coffee plantations. This process was led by the Laboratorio de Pesticidas of Universidad del Quindío (Colombia). Besides, a negative control group of thirty volunteers who had not been exposed to pesticides was included.
All the participants signed an informed consent (previously approved by the ethical committee) to take part in the study. The inclusion criteria included male subjects aged 18 or older and living in the Colombian Coffee Region.
A blood sample was taken from each participant and processed to obtain blood plasma. Each plasma was analyzed to evaluate the presence and concentration of pesticides using Gas Chromatography with Flame Ionization Detector (GC-FID). Out of the 100 cases, 27 were found to be below the detection limit and considered negative cases, while 73 were found to be above the detection limit and considered positive cases.
In the plasma of the 73 positive cases, the presence of six organochlorine pesticides (i.e., endosulfan, endrin, heptachlor, DDT, methoxychlor, and lindane) and chlorpyrifos (an organophosphorus pesticide) was identified. Furthermore, to assess the metabolic profile, the samples were processed and derivatized following the same protocol used for cell extracts [
2.3. Statistical analysis
PLS-DA and SVM-RFE were performed here to evaluate the metabolic profiles taken from the in vitro and in vivo studies. In the in vitro data, a subset with 153 metabolites was identified. Five groups were defined, one for each organochlorine: aldrin, DDT, endosulfan, lindane, and the mixture. Each group was compared with the control; hence, each test consisted of six experimental replicates.
Regarding the in vivo data, the plasma samples were classified into seven groups according to the pesticide found in them: 5 samples in endosulfan, 31 in endrin, 28 in heptachlor, 3 in DDT, 4 in methoxychlor, 35 in lindane, and 18 in chlorpyrifos. Each group was then compared with the negative control.
MetaboAnalyst 4.0 was used to perform PLS-DA [
The list of the ten most relevant metabolites was obtained compiled using the Variable Importance in Projection (VIP) score [
Incremental training was carried out with 20 %, 40 %, 60 %, and 80 % of the samples in order to identify the lowest number of samples needed to achieve 100 % accuracy (measured by k-fold cross-validation). SVM training was conducted with both normalized and raw data. The kernel with the best performance and minimum sample size required for training was employed to implement the SVM-RFE algorithm, but, in this case, using 100 % of the available samples. The lists with the ten most relevant metabolites were obtained for each metabolic profile.
2.4. Comparative analysis
The two methods were compared based on the accuracy results of the k-fold cross-validation and the position and inclusion of metabolites in the lists obtained by both.
3. RESULTS
Data normalization with log transformation, Pareto scaling, and PLS-DA were performed using MetaboAnalyst 4.0. PLS-DA was conducted with normalized data.
Regarding PLS-DA, although accuracy was measured by k-fold cross-validation, it should be noted that MetaboAnalyst 4.0 requires a minimum of ten samples to apply such method. This criterion was not fulfilled by the DDT and methoxychlor samples in the in vivo study. In those cases, accuracy was validated using the LOOCV algorithm. Note that the results shown in Figure 1 represent the first principal component (the component with the best score).
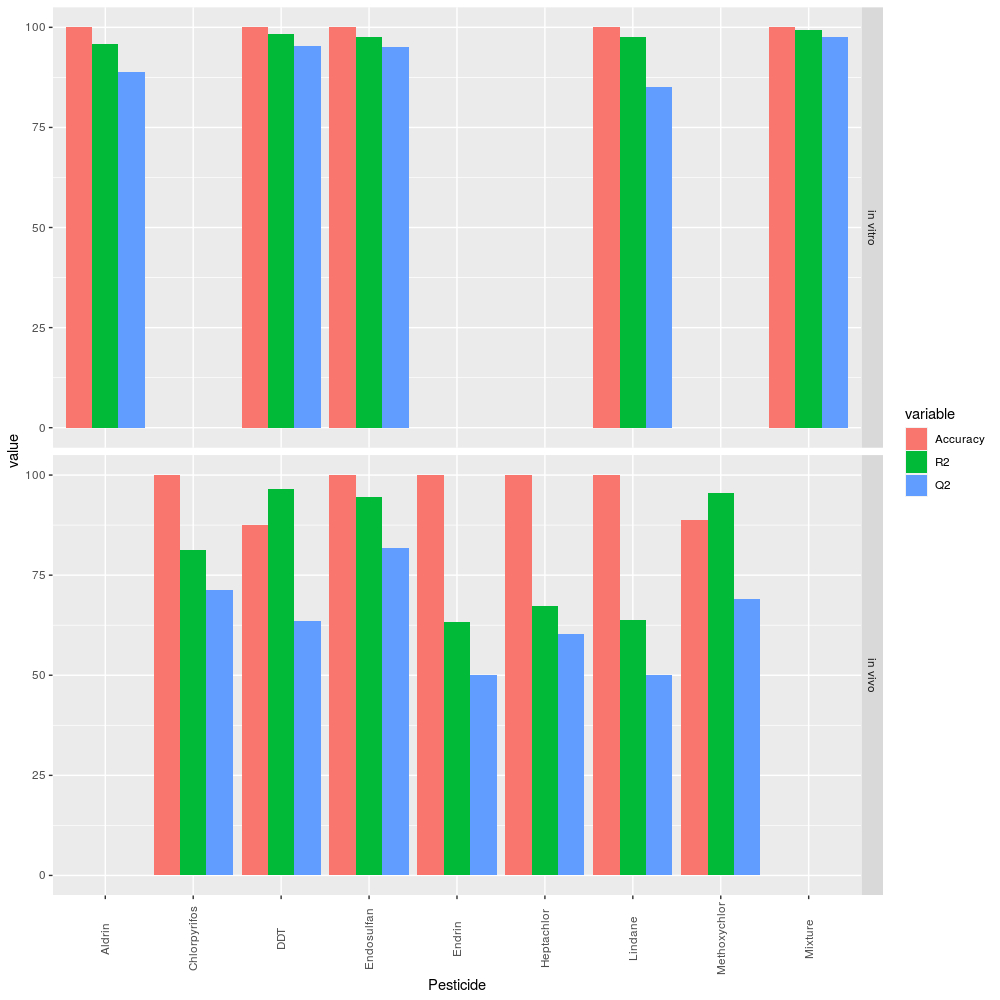
From Figure 1, we observe that, when PLS-DA was implemented using the data from the in vitro study, R2 was above 95 %; and Q2, above 85 %.
However, when implemented using the data from the in vivo study, its accuracy decreased in those groups in which there were fewer samples. R2 fell to 63.4 % (endrin). In addition, Q2 was also affected; it fell to 50.05 % (lindane) and did not exceed 81.9 % (endosulfan).
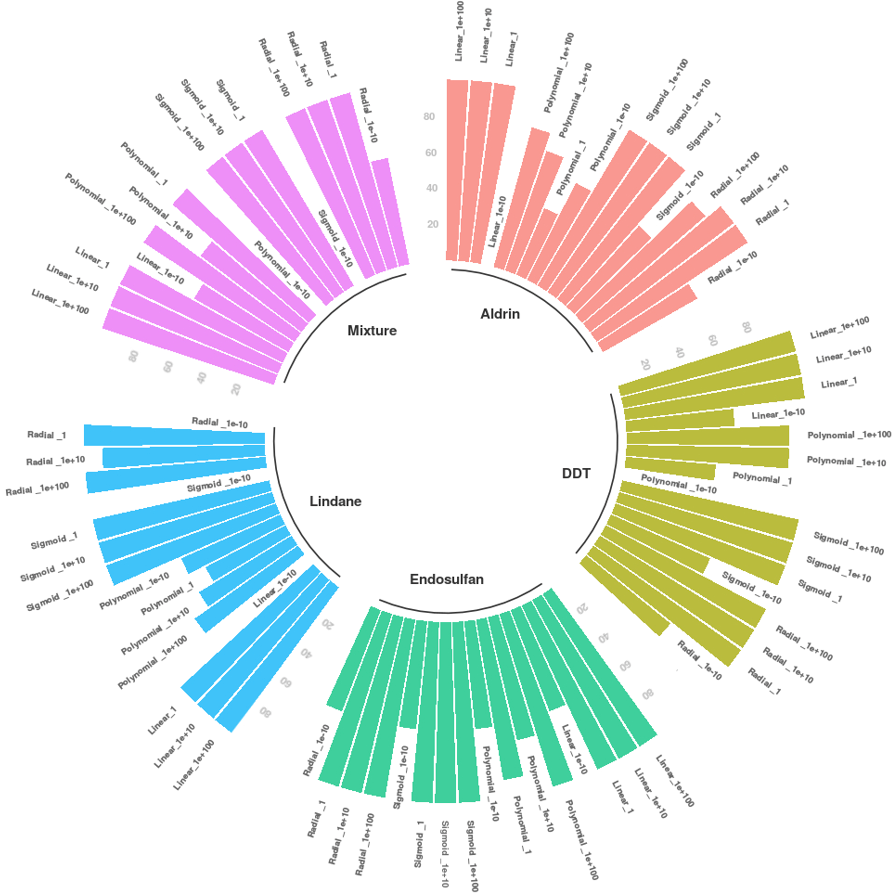
SVMs were applied to both normalized and raw data. Nevertheless, the best results were achieved with normalized data; they are shown in Figures 2 and 3. In these, SVM training was performed with 80 % of the data, and 20 % was used to test the SVM model obtained. Four kernels were evaluated in terms of SVM training: Linear, Polynomial, Sigmoid, and Radial. Four cost margin penalties were implemented in each kernel: 1e100, 1e10, 1, and 1e-10. Kernel and cost used in each model are specified in the figure. Bar sizes represent the prediction accuracy obtained by each SVM model in a scale between 0 % and 100 %.
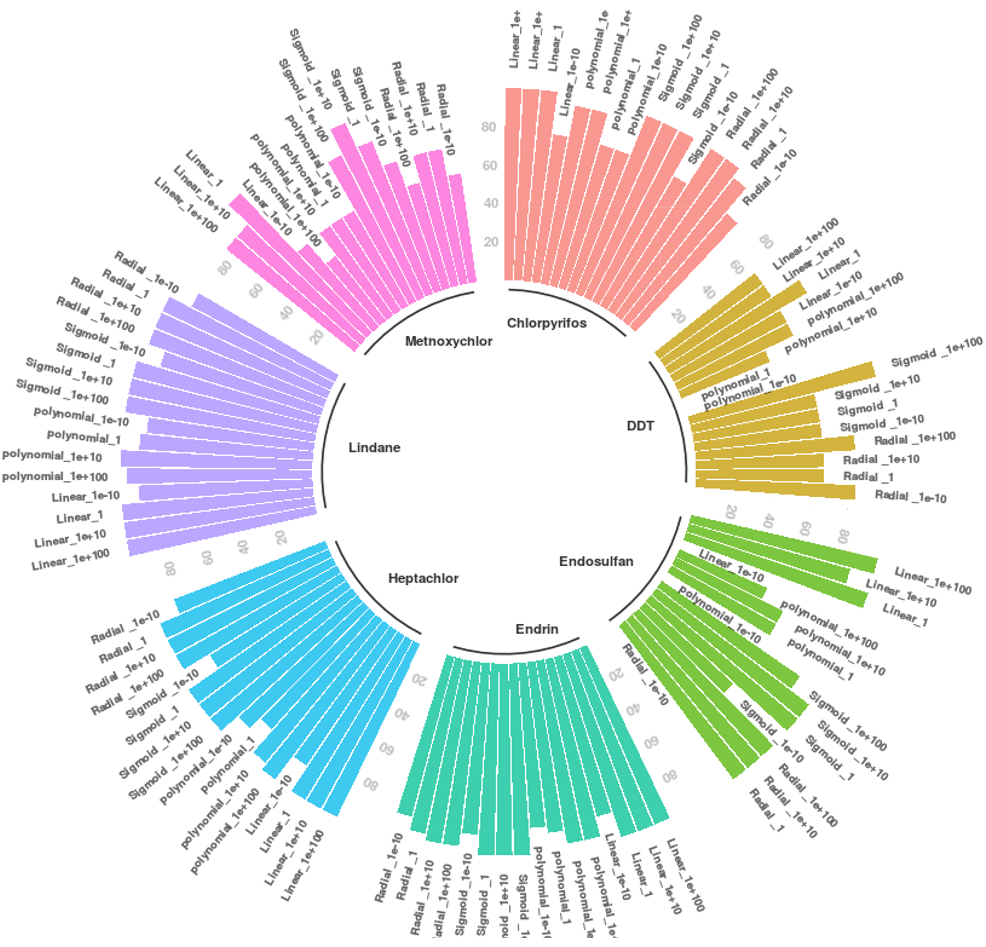
Employing the normalized data from the in vitro study, all kernels exhibited good performance (except for the polynomial one) with 100 % accuracy using 80 % of the samples for training. Implementing the normalized data from the in vivo study, there was a slight decrease in accuracy, especially in those groups with a number of samples below ten (DDT and methoxychlor). However, the latter achieved 100 % accuracy in some scenarios of the sigmoid kernel.
With respect to raw data, the best performance was achieved using 80 % of the samples for training, and the accuracy was between 90.63 % and 93.15 %. In addition to the linear kernel, the polynomial and radial kernels showed good performance. The polynomial kernel in particular yielded an accuracy of 93.1 % using a margin penalty of 1e-10. This case opens up the possibility of overfitting.
In PLS-DA, the relevant features were identified by means of VIP scores, while, in SVM, the SVM-RFE technique was employed for such purpose. Both scenarios used normalized data. Tables 1 and 2 show the features proposed by both methods for the in vitro study.
| Aldrin | DDT | Endosulfan | Lindane | Mixture |
| Phosphoethanolamine | Phosphoethanolamine | N-acetyl aspartate | Taurine | Glucose-6-phosphate |
| Phosphogluconic acid | Cytidine-5 -monophosphate | Citric acid | Phosphogluconic acid | Phosphoethanolamine |
| Cytosin | Phosphogluconic acid | Taurine | Gluconic acid | Citric acid |
| Cysteine | Glutathione | Glucose-6-phosphate | Alpha-ketoglutarate | Isocitric acid |
| Gluconic acid | 2.5-dihydroxy pyrazine | Phosphogluconic acid | N-acetyl mannosamine | Phosphogluconic acid |
| Ribulose-5-phosphate | 5'-deoxy-5'-methyl thio adenosine | Phosphoethanolamine | Glutaric acid | Ribose |
| Hypoxanthine | Cytosin | Alpha-keto glutarate | 2.5-dihydroxy pyrazine | Aspartic acid |
| Alpha ketoglutarate | Gluconic acid | Isocitric acid | Alpha-amino adipic acid | Hypoxanthine |
| Fructose 1 phosphate | Hexitol | Hexose-6-phosphate | Adenine | Hexose-6-phosphate |
| Aspartic acid | Sulfuric acid | Cysteine | Ribulose-5-phosphate | Alpha-ketoglutarate |
| Aldrin | DDT | Endosulfan | Lindane | Mixture |
| Phosphogluconic acid | Phosphoethanolamine | Citric acid | Alpha ketoglutarate | Glucose 6 phosphate |
| Alpha aminoadipic acid | Cytosin | Phosphogluconic acid | Phosphogluconic acid | Phosphogluconic acid |
| Phosphoethanolamine | 2,5 dihydroxypyrazine | Isocitric acid | Taurine | Phosphoethanolamine |
| Cysteine | Alpha aminoadipic acid | Alpha ketoglutarate | N acetylmannosamine | Citric acid |
| Gluconic acid | Phosphogluconic acid | Hexose 6 phosphate | Glycerol alpha phosphate | Ribose |
| Cytosin | Gluconic acid | Aspartic acid | Gluconic acid | Cytidine 5 monophosphate |
| Hypoxanthine | Aspartic acid | N acetylaspartate | Xilitol | Isocitric acid |
| Alpha ketoglutarate | Cytidine 5 monophosphate | Hypoxanthine | Creatinine | Hypoxanthine |
| 3 phosphoglycerate | Ribulose 5 phosphate | 3 phosphoglycerate | 2 5 dihydroxypyrazine | Alpha ketoglutarate |
| Malic acid | Glutathione | Aconitic acid | Asparagine | Cysteine |
The results of the in vivo study are not reported because no identification of the compounds was performed in that case. However, the percentage of coincidence between the two methods (i.e., PLS-DA and SVM-RFE) in the two studies was calculated here (Table 3).
| Data from the in vitro study | ||
| Pesticide | Included in the top ten | Same position in the top |
| Aldrin | 70.00 | 40.00 |
| DDT | 60.00 | 10.00 |
| Endosulfan | 60.00 | 0.00 |
| Lindane | 60.00 | 10.00 |
| Mixture | 80.00 | 20.00 |
| Data from the in vivo study | ||
| Pesticide | Included in the top ten | Same position in the top |
| Chlorpyrifos | 50.00 | 20.00 |
| DDT | 30.00 | 0.00 |
| Endosulfan | 50.00 | 0.00 |
| Endrin | 70.00 | 0.00 |
| Heptachlor | 80.00 | 20.00 |
| Lindane | 80.00 | 10.00 |
| Methoxychlor | 60.00 | 10.00 |
The comparative analysis reveals that, in the in vitro study, aldrin and the mixture were found to have the highest coincidences among the relevant metabolites identified in each method. Nevertheless, this panorama changes in the in vivo study, as heptachlor and lindane exhibited the highest number of coincidences.
4. DISCUSSION
In this study, PLS-DA was proven to be a good method to analyze data from in vitro studies, as it presented an R2 and a Q2 close to ideal values. However, when analyzing data from in vivo studies, its accuracy decreased in scenarios with few samples. Conversely, SVMs achieved 100 % accuracy in all the scenarios (in vitro and in vivo), but it was necessary to test the performance of the different kernels. Although the linear and sigmoid kernels exhibited good performance using margin penalties of 1e100, 1e10, and 1, the radial and polynomial kernels showed a poor one.
According to this, the accuracy of PLS-DA and SVMs can be affected by conditions such as high variability and few samples, like those in in vivo studies. Nonetheless, it is possible to identify the kernels with the best performance for data analysis from in vivo studies and use them in SVMs, thus allowing a better classification. Moreover, another advantage of SVMs is that they can achieve an accuracy of 100 % with fewer training samples. For instance, in this study, they employed 80 % of the samples, while PLS-DA required all of them.
Furthermore, comparing the lists of the ten relevant features of each profile in each method (SVM-RFE and PLS-DA), both methods shared similarities in the analysis of the in vitro study (equal to or greater than 70 %), but the results were heterogeneous for the in vivo study. The group with the lowest number of coincidences was DDT in the in vivo study, which poses the question of whether the number of samples could have influenced these results.
This study identified an improvement in the predictive performance of SVMs over PLS-DA in the analysis of data from in vivo experiments, something previously described by Mahadevan et al.[
In this study, the linear and sigmoid kernels showed the best performance. Although the radial kernel did not exhibit an adequate performance in this article, it has been one of the most widely employed [
Although this study focused on the classic SVM kernels, new kernels have been proposed, such as the Hermite orthogonal polynomial kernel. This kernel makes it possible to use fewer support vectors for classification. In addition, it has been reported to achieve better error-rate performance [
SVM-RFE was employed here to select a list of relevant features. For this purpose, we suggest implementing SVMs with a kernel having an optimal margin penalty before using SVM-RFE. In particular, in this study, the linear and sigmoid kernels presented a margin penalty that was optimal for most scenarios. Nevertheless, for scenarios with few samples such as DDT, the sigmoid kernel was the only one that showed optimal performance. Although there were enough samples for data comparison in the in vitro study, some scenarios in the in vivo study, such as DDT and methoxychlor, had few samples. In this case, there was the risk of overfitting in both methods.
Furthermore, it should be noted that, in the in vivo study, among the 73 cases with proven pesticide exposure, some agricultural workers had been exposed to more than one pesticide, which could have influenced the metabolic profiles and, hence, the performance of each technique.
Although we identified 153 metabolites from the spectrometry signals obtained in the in vitro experiment, this was not done in the in vivo study, but it remains to be performed in order to define the biological impact in each scenario.
In addition to SVM-RFE, another strategy that has been proposed to identify relevant features is multiclass SVM using L1-norm [
In summary, according to the findings of this work and those of the other studies mentioned here, SVMs are robust methods suitable for data derived from in vivo experiments and exhibit good classification performance even with few samples. Also, SVMs are advantageous in dealing with outliers, predictive power, and resistance to overfitting. However, their performance will depend on the hyperparameters and kernels used.
Therefore, in order to make the most of the analysis with SVM-RFE and the “kernel trick”, it is recommended to initially evaluate each kernel, as well as the different margin penalty scenarios. Performance must also be evaluated based on the percentage of samples used for training in order to avoid overfitting. In this study, 80 % of the samples were needed for most scenarios. However, this may vary depending on the number of features and samples available. Next, the last step would be to implement SVM-RFE with the best kernel identified.
Finally, it is necessary to clarify that the results obtained from one or the other method should be validated in future biological experiments to determine the biological impact of exposure to pesticides.
5. CONCLUSIONS
In this study, SVMs and PLS-DA were proven to be appropriate methods to analyze data from in vitro studies with controlled conditions, but PLS-DA presented difficulties with data from in vivo studies (non-controlled conditions and non-linear data) in the context of organochlorine exposure.
Regarding class prediction in data from in vivo studies, SVMs exhibited a greater predictive power than PLS-DA. Moreover, the kernel with the best performance identified by SVM analysis can be used in SVM-RFE to obtain an adequate list of most relevant features in the context of pesticides exposure. Additionally, the computational cost of SVMs is low.
SVM-RFE is becoming a useful tool for biomarker identification, even when there are few samples. In addition, it is considered a robust method to analyze data derived from in vivo and in vitro studies.
6. ACKNOWLEDGMENTS
The authors wish to thank the B2SLab of the Universitat Politècnica de Catalunya for providing the equipment to conduct the analyses, the Instituto Tecnológico Metropolitano (ITM) for funding the exchange program that led to the results of this study, and ITM Language Center for proofreading the manuscript.
CONFLICTS OF INTEREST
All authors declare that there were no conflicts of financial, professional, or personal interest that could inappropriately influence the results obtained in the present study.
AUTHOR CONTRIBUTIONS
Jorge Alejandro Lopera-Rodríguez contributed with developing the research idea, conducting the analysis using SVM-RFE and PLS-DA, implementing the methods in R, carrying out the comparative analysis, writing the article (introduction, methods, results, discussion, conclusions, and tables), and translating it.
Martha Zuluaga contributed with conducting the in vitro and in vivo experiments, designing the figures, writing the article (methods, results, and discussion), and translating it.
Jorge Alberto Jaramillo-Garzón contributed with assessing the analysis methods using PLS-DA and SVM-RFE, supervising the application of these methods, and supervising and correcting the article.
7. REFERENCES
- arrow_upward [1] J. C. Lindon; J. K. Nicholson: E. Holmes, The Handbook of Metabonomics and Metabolomics. Elsevier, 2007.
- arrow_upward [2] E. C. Horning; M. G. Horning, “Human Metabolic Profiles Obtained by GC and GC/MS,” J. Chromatogr. Sci., vol. 9, no. 3, pp. 129–140, Mar. 1971. https://doi.org/10.1093/chromsci/9.3.129
- arrow_upward [3] S. Mahadevan; S. L. Shah; T. J. Marrie; C. M. Slupsky, “Analysis of Metabolomic Data Using Support Vector Machines,” Anal. Chem., vol. 80, no. 19, pp. 7562–7570, Sep. 2008. https://doi.org/10.1021/ac800954c
- arrow_upward [4] C. Cortes; V. Vapnik, “Support-vector networks,” Mach. Learn., vol. 20, no. 3, pp. 273–297, Sep. 1995. https://doi.org/10.1007/BF00994018
- arrow_upward [5] A. Alonso; S. Marsal; A. JuliÃ, “Analytical Methods in Untargeted Metabolomics: State of the Art in 2015,” Front. Bioeng. Biotechnol., vol. 3, p. 23, Mar. 2015. https://doi.org/10.3389/fbioe.2015.00023
- arrow_upward [6] J. Heinemann; A. Mazurie; M. Tokmina-Lukaszewska; G. J. Beilman; B. Bothner, “Application of support vector machines to metabolomics experiments with limited replicates,” Metabolomics, vol. 10, no. 6, pp. 1121–1128, Dec. 2014, https://doi.org/10.1007/s11306-014-0651-0
- arrow_upward [7] K. M. Mendez; S. N. Reinke; D. I. Broadhurst, “A comparative evaluation of the generalised predictive ability of eight machine learning algorithms across ten clinical metabolomics data sets for binary classification,” Metabolomics, vol. 15, no. 12, p. 150, Nov. 2019. https://doi.org/10.1007/s11306-019-1612-4
- arrow_upward [8] P. S. Gromski et al., “A tutorial review: Metabolomics and partial least squares-discriminant analysis – a marriage of convenience or a shotgun wedding,” Anal. Chim. Acta, vol. 879, pp. 10–23, Jun. 2015. https://doi.org/10.1016/j.aca.2015.02.012
- arrow_upward [9] I. Guyon; J. Weston; S. Barnhill; V. Vapnik, “Gene selection for cancer classification using support vector machines,” Mach. Learn., vol. 46, no. 1, pp. 389–422, Jan. 2002. https://doi.org/10.1023/A:1012487302797
- arrow_upward [10] W. Guan et al., “Ovarian cancer detection from metabolomic liquid chromatography/mass spectrometry data by support vector machines,” BMC Bioinformatics, vol. 10, no. 259, Aug. 2009. https://doi.org/10.1186/1471-2105-10-259
- arrow_upward [11] X. Lin et al., “A support vector machine-recursive feature elimination feature selection method based on artificial contrast variables and mutual information,” J. Chromatogr. B, vol. 910, pp. 149–155, Dec. 2012. https://doi.org/10.1016/j.jchromb.2012.05.020
- arrow_upward [12] M. Abdollahi; A. Ranjbar; S. Shadnia; S. Nikfar; A. Rezaiee, “Pesticides and oxidative stress: a review,” Med. Sci. Monit., vol. 10, no. 6, Jun. 2004. https://pubmed.ncbi.nlm.nih.gov/15173684/
- arrow_upward [13] V. Moses; J. V. Peter, “Acute intentional toxicity: endosulfan and other organochlorines,” Clin. Toxicol., vol. 48, no. 6, pp. 539–544, Jul. 2010. https://doi.org/10.3109/15563650.2010.494610
- arrow_upward [14] R. Jayaraj; P. Megha; P. Sreedev, “Organochlorine pesticides, their toxic effects on living organisms and their fate in the environment,” Interdiscip. Toxicol., vol. 9, no. 3–4, p. 90- 100, Dec. 2016. https://doi.org/10.1515/intox-2016-0012
- arrow_upward [15] M. Zuluaga; J. J. Melchor; F. A. Tabares-Villa; G. Taborda; J. C. Sepúlveda-Arias, “Metabolite Profiling to Monitor Organochlorine Pesticide Exposure in HepG2 Cell Culture,” Chromatographia, vol. 79, no. 17–18, pp. 1061–1068, Sep. 2016. https://doi.org/10.1007/s10337-016-3031-2
- arrow_upward [16] O. Fiehn; T. Kind, “Metabolite Profiling in Blood Plasma,” in Metabolomics, Springer, 2007, pp. 3–17. https://doi.org/10.1007/978-1-59745-244-1_1
- arrow_upward [17] O. Fiehn et al., “Quality control for plant metabolomics: reporting MSI-compliant studies,” Plant J., vol. 53, no. 4, pp. 691–704, Feb. 2008. https://doi.org/10.1111/j.1365-313X.2007.03387.x
- arrow_upward [18] J. Chong; D. S. Wishart; J. Xia, “Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis,” Curr. Protoc. Bioinforma., vol. 68, no. 1, p. e86, Sep. 2019. https://doi.org/10.1002/cpbi.86
- arrow_upward [19] L. Eriksson, Introduction to multi-and megavariate data analysis using projection methods (PCA & PLS). Umetrics AB, 1999.
- arrow_upward [20] R. C. Team, “R: A language and environment for statistical computing,” 2013. https://www.yumpu.com/en/document/read/6853895/r-a-language-and-environment-for-statistical-computing
- arrow_upward [21] M. Campbell, “RStudio Projects,” in Learn RStudio IDE, Berkeley, CA: Apress, 2019, pp. 39–48. https://doi.org/10.1007/978-1-4842-4511-8_4
- arrow_upward [22] D. Meyer et al., “Package ‘e1071, Misc Functions of the Department of Statistics, Probability Theory Group (Formerly: E1071), TU Wien’”, versió 1.7-9, R J., 2019. http://sunsite2.icm.edu.pl/pub/unix/math/cran/web/packages/e1071/e1071.pdf
- arrow_upward [23] H. Zheng et al., “Predictive diagnosis of major depression using NMR-based metabolomics and least-squares support vector machine,” Clin. Chim. Acta, vol. 464, pp. 223–227, Jan. 2017. https://doi.org/10.1016/j.cca.2016.11.039
- arrow_upward [24] B. Feizizadeh; M. S. Roodposhti; T. Blaschke; J. Aryal, “Comparing GIS-based support vector machine kernel functions for landslide susceptibility mapping,” Arab. J. Geosci., vol. 10, no. 122, Mar. 2017. https://doi.org/10.1007/s12517-017-2918-z
- arrow_upward [25] M. A. Horaira; M. S. Ahmed; M. H. Kabir; M. N. H. Mollah; M. A. Rahman Shah, “Colon Cancer Prediction from Gene Expression Profiles Using Kernel Based Support Vector Machine,” in 2018 International Conference on Computer, Communication, Chemical, Material and Electronic Engineering (IC4ME2), Feb. 2018, pp. 1–4. https://doi.org/ 10.1109/IC4ME2.2018.8465636
- arrow_upward [26] V. Wan; W. M. Campbell, “Support vector machines for speaker verification and identification,” in Neural Networks for Signal Processing X. Proceedings of the 2000 IEEE Signal Processing Society Workshop (Cat. No.00TH8501), vol. 2, pp. 775–784. https://doi.org/10.1109/NNSP.2000.890157
- arrow_upward [27] V. Hooshmand Moghaddam; J. Hamidzadeh, “New Hermite orthogonal polynomial kernel and combined kernels in Support Vector Machine classifier,” Pattern Recognit., vol. 60, pp. 921–935, Dec. 2016. https://doi.org/10.1016/j.patcog.2016.07.004
- arrow_upward [28] X. Huang; Q.-S. Xu; Y.-H. Yun; J.-H. Huang; Y.-Z. Liang, “Weighted variable kernel support vector machine classifier for metabolomics data analysis,” Chemom. Intell. Lab. Syst., vol. 146, pp. 365–370, Aug. 2015. https://doi.org/10.1016/j.chemolab.2015.06.009
- arrow_upward [29] D. A. López-Sarmiento; H. C. Manta-Caro; N. E. Vera-Parra, “Clasificador basado en una máquina de vectores de soporte de mínimos cuadrados frente a un clasificador por regresión logística ante el reconocimiento de dígitos numéricos,” TecnoLógicas, no. 31, pp. 37-51, Nov. 2011. https://doi.org/10.22430/22565337.99
- arrow_upward [30] L. A. Muñoz-Bedoya; L. E. Mendoza; H. J. Velandia-Villamizar, “Segmentación de Imágenes de Resonancia Magnética IRM utilizando LS-SVM y Análisis Multiresolución Wavelet,” TecnoLógicas, pp. 681-693, Nov. 2013. https://doi.org/10.22430/22565337.381
- arrow_upward [31] M. Moon; K. Nakai, “Stable feature selection based on the ensemble L 1 -norm support vector machine for biomarker discovery,” BMC Genomics, vol. 17, no. s13, Dec. 2016. https://doi.org/10.1186/s12864-016-3320-z